January 28, 2025
Sonya Dyhrman (Lamont-Doherty Earth Observatory, Columbia Univ.) - Learning the rules of the system: Tracking drivers of microeukaryote physiological ecology in the North Pacific Subtropical Gyre
Lucia Campese (Stazione Zoologica Anton Dohrn, Naples) - NEREA Initiative: Integrating Multi-Omics Approaches for Marine Observations in Napless
August 27, 2024
Sarah Smith (Moss Landing Marine Laboratories) - Metatranscriptomics of Monterey Bay Phytoplankton During Experimental Upwelling Mesocosm Experiments
Mora Groussman (Univ. Washington) - Using the MarFERReT eukaryotic sequence library for dynamic, custom and reproducible reference libraries
May 1, 2024: theme: Omics intercomparisons and proteomic/transcriptomic applications in the field
Mak Saito (WHOI) - Intercomparison of ocean metaproteomics analysis: Initial results of global data-dependent acquisition analyses
Erin Bertrand (Dalhousie University) - Toward understanding rates of eukaryotic microbial processes through metaproteomics
Microeukaryotes are core components of the biological carbon pump through their roles in primary production, carbon export, and trophic transfer in the marine food web.
Metagenomics, metatranscriptomics, metabarcoding, and eDNA sampling are increasingly being performed as core measurements on individual cruises as well as being incorporated into large oceanographic surveys (e.g. Tara Oceans, Bio-GO-SHIP, BioGeoTraces). However, there is currently no single gold standard practice for the collection, preservation, or processing of these samples—nor is there a sense of how variable these measures might be among labs or research cruise endeavors. Intercomparison and intercalibration of ‘omic studies is a necessary next step for our field.
The goal of this activity is to determine the extent of variability in existing metatranscriptomic pipelines through a deliberate community-led intercomparison to build international confidence in methodological choices.
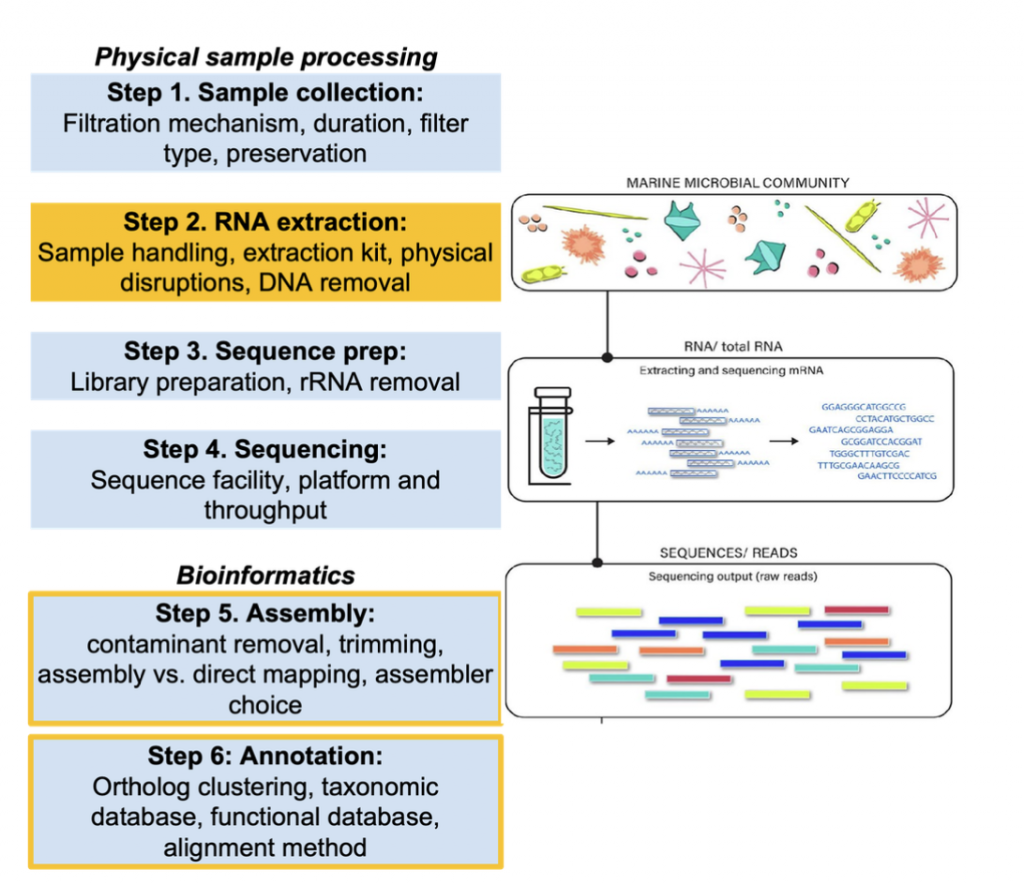
Figure caption: Overview of the metatranscriptomic collection and analysis process. This intercalibration exercise focuses on the steps highlighted in orange (extraction and bioinformatic analyses). Briefly, each participant was assigned at least two filter slices that will be used to test various comparisons. The intercomparison will begin at Step 2: RNA extraction, given that samples have already been collected using in-situ McLane pumps in Summer 2023. After sequences are obtained, all samples will be pooled and used with a consistent pipeline to determine the effects of Steps 1-4. In addition, users will be asked to process a designated sample using their own computational procedure to isolate sources of variation in the bioinformatic steps (Steps 5 & 6)
Planned Activities & Products:
Capacity building
For in person meetings 10 early career researchers (late graduate students and postdoc scholars) will be selected from open applications – (criteria: graduate program of study and applicability to the intercomparison, interest level in bioinformatics, and career stage).
EC researchers will gain advanced computational biology training in the first meeting; EC ideas and intercomparison results will be included in the planned publication via their participation in the second meeting.