This summer OCB and GEOTRACES are co-sponsoring a synthesis workshop on the biogeochemical cycling of trace elements in the ocean. The overall goal of the workshop is to bring together expertise from across the field of oceanography to take advantage of the growing datasets of trace elements in the ocean and explore biological-chemical-physical underpinnings of trace element cycling within the ocean. One of the three main themes that will be addressed at the workshop is “biological uptake and trace element bioavailability.” Part of this theme will include a discussion of how molecular markers have been used to address questions of trace element bioavailability in the past and the exciting future for continued efforts in this area given the growing molecular toolkit.
As a starting point to open the door to this broad discussion topic, let us turn our attention to how molecular methods have been used to evaluate iron (Fe) bioavailability to certain phytoplankton groups. Marine phytoplankton play a key role in the global carbon cycle by performing a significant fraction of global primary production (1). Since John Martin’s groundbreaking work introduced the concept that Fe is a limiting nutrient for phytoplankton growth (2), numerous studies have shown that insufficient Fe limits primary productivity in the major high-nitrate, low-chlorophyll (HNLC) regions of the ocean, and that Fe availability can regulate phytoplankton processes in many other oceanic settings (3, 4). Fe is a critical micronutrient required by phytoplankton for a Exploring molecular methods for assessing trace element bioavailability in phytoplankton Dreux Chappell (Old Dominion Univ.) multitude of cellular tasks, including electron transfer in photosynthesis and respiration, as well as macronutrient acquisition and assimilation (5). Studies of Fe limitation in marine environments beyond the traditional HNLC regions suggest that Fe limitation may be driven not simply by low Fe concentrations, but a combination of low Fe bioavailability coupled with high macronutrient supply (e.g., 6, 7). In an admittedly overly simplified summary, the different pools of Fe present in the ocean are defined based on filter pore-size cut-offs and chemical interactions with organic compounds (3). Knowledge of the distribution of these various forms of Fe in the oceans has increased dramatically in recent years thanks to coordinated sampling efforts like GEOTRACES.
While the bioavailability of Fe to phytoplankton is believed to be different for the various Fe pools, there is still no clear consensus as to which, if any, pools of Fe are always bioavailable and which, if any, are completely unavailable (8, 9). Additionally, it is known that all phytoplankton are not created equal with respect to their ability to persist under low-Fe conditions (10) and access different Fe pools (9). These factors make it difficult to accurately predict how changing concentrations of the different pools of Fe may impact phytoplankton productivity in a changing ocean. One way to address questions about biological availability is to use phytoplankton themselves as in situ indicators of Fe stress. An approach that appears promising in addressing these questions of bioavailability of Fe to individual phytoplankton species is the development of species-specific molecular markers of Fe limitation such as those that have been developed for the oceanic diatom Thalassiosira oceanica (11), and for the two main groups of the nitrogen-fixing cyanobacterial genus Trichodesmium (12). These assays in particular follow the expression of genes that encode flavodoxin, a non-Fe-containing protein that phytoplankton are known to substitute for the Fe-containing ferrodoxin protein to maintain photosynthetic electron transport under Fe-limiting conditions (13). They focus on gene expression analysis rather than protein analysis, as many phytoplankton have multiple genes that encode for flavodoxin proteins and, at least in diatoms, not all gene copies are sensitive to Fe (14).
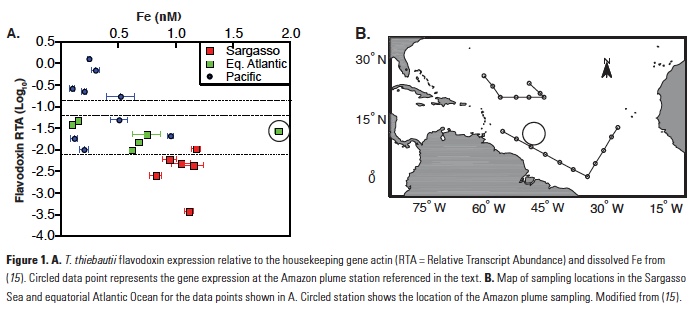
The Trichodesmium assays were calibrated using cultures grown with six different concentrations of Fe in the media. In laboratory cultivation experiments, gene expression was shown to be inversely proportional to Fe present in the media and expression was downregulated when Fe was fed back to Fe-limited cultures (12). The Trichodesmium thiebautii assay was further used to evaluate field populations from open ocean samples collected globally (15), providing insights into Fe bioavailability to wild populations of Trichodesmium. Overall, there was an inverse correlation between gene expression and total dissolved Fe concentrations (Fig. 1). Comparing the results with the laboratory calibration led to the conclusion that most of the dissolved Fe, including organically bound Fe, was available to T. thiebautii. An intriguing result from this study was that one sample collected in the plume of the Amazon River had significantly higher gene expression than would be expected based on the measured dissolved Fe at that site (Fig. 1; 15). These findings suggest that there is a fraction of the dissolved Fe in the Amazon River plume that is not bioavailable to T. thiebautii, the clade of Trichodesmium that is more abundant and active in the open ocean (15, 16).
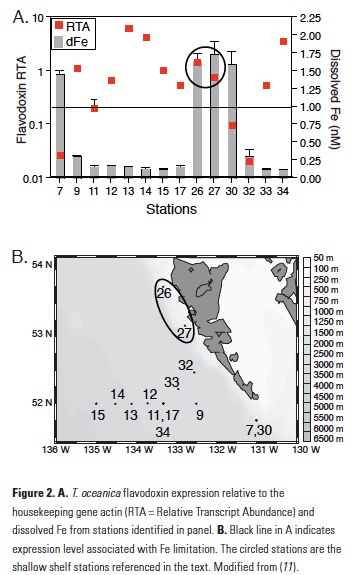
The T. oceanica assay is also highly sensitive to Fe with high gene expression in cultures that were Fe-limited, a rapid reduction of gene expression following an Fe pulse to Fe-limited cultures, and no induction of expression by macronutrient limitation (11). Using this method on field samples from the northeast Pacific Ocean, T. oceanica flavodoxin expression was found to be highest in samples with low measured dissolve Fe and vice versa (11). Two notable exceptions to this trend were samples collected along the shallow shelf of Haida Gwaii, stations 26 and 27, which showed anomalously high expression of both genes despite high measured dissolved Fe (Fig. 2), suggesting that something about the dissolved Fe in these shallow coastal stations made it unavailable to the T. oceanica in these waters. It should be noted that T. oceanica is an oceanic diatom species, so both sets of findings suggest that there is a fraction of dissolved Fe from a terrestrially influenced water sample that was not bioavailable to a species of oceanic phytoplankton.
While these two datasets are intriguing, they are limited in scope and admittedly raise more questions than they answer. There are a variety of questions that stem from these results, including what, if anything, is different about the dissolved Fe at these stations? Was this coastal/terrestrially sourced Fe unavailable because only oceanic phytoplankton, which rarely encounter dissolved Fe from terrestrial sources, were queried? Did coastal phytoplankton simply outcompete the oceanic phytoplankton for access to the dissolved Fe perhaps because they have different or more efficient Fe uptake mechanisms? Would an assay targeting coastal phytoplankton reveal the same results? We know that coastal diatoms have significantly higher Fe requirements (10). What if coastal and oceanic phytoplankton also differ in their abilities to access Fe from different sources? A recent study in the Sea of Okhotsk yielded a significant correlation between a bulk diatom community indicator of Fe stress and dissolved Fe with increasing distance from the mouth of the Amur River, suggesting that the riverine Fe was bioavailable to the community that was dominated by coastal diatoms (17).
A number of recent advances in molecular microbial oceanography are making it increasingly possible to start answering these types of questions on a broader scale. A major sequencing effort that was completed in 2014, the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP), added a wealth of data on the functional genetic diversity of marine microeukaryotes (18). The project resulted in over 650 publicly available transcriptomes from over 250 genera of marine microeukaryotes. Even before all the transcriptomes from the project were released, data mining of a limited portion of the MMETSP publically available dataset yielded valuable information as to the varied Fe management strategies utilized by different species of marine diatoms, creating a list of additional potential molecular markers for evaluating Fe nutritional status of diatoms in the field (19). Another data mining technique that has proven useful in identifying coordinated transcriptional responses in diatoms has involved the application of clustering algorithms to evaluate publically available microarray data for two of the more commonly studied diatom species, Phaeodactylum tricornutum and Thalassiosira pseudonana, grown under a wide variety of conditions (20). New insights into the diatom Fe stress response and Fe uptake mechanisms have also been gained through combining physiological experiments with genetic knockdowns of previously uncharacterized Fe responsive genes (21). These new molecular advances are providing a suite of new potential targets for querying the physiological status of phytoplankton present throughout the global ocean. At the OCB/GEOTRACES synthesis workshop on the biogeochemical cycling of trace elements in the ocean, the conversation will include a discussion of how to effectively combine these new analyses with the growing datasets of bioactive trace elements to answer questions regarding the biological availability of different trace element pools. While this mini-review has focused on Fe, additional trace elements will be discussed at the meeting in August.
Author
Dreux Chappell (Old Dominion University)
References
1. C. B. Field et al., Science 281, 237-240 (1998).
2. J. H. Martin, S. Fitzwater, Nature 331, 341-343 (1988).
3. P. W. Boyd, M. J. Ellwood, Nature Geoscience 3, 675-682 (2010).
4. P. W. Boyd et al., Science 315, 612-617 (2007).
5. F. M. M. Morel, N. M. Price, Science 300, 944-947 (2003).
6. K. W. Bruland, E. L. Rue, G. J. Smith, Limnol. Oceanogr. 46, 1661-1674 (2001).
7. A. L. King, K. N. Buck, K. A. Barbeau, Marine Chem. 128-129, 1-12 (2012).
8. E. Breitbarth et al., Biogeosci. 7, 1075-1097 (2010).
9. Y. Shaked, H. Lis, Frontiers Microbiol. 3 (2012).
10. W. G. Sunda, S. A. Huntsman, Marine Chem. 50, 189-206 (1995).
11. P. D. Chappell et al., ISME Journal 9, 592-602 (2015).
12. P. D. Chappell, E. A. Webb, Envir. Microbiol. 12, 13-27 (2010).
13. J. LaRoche et al., J. Phycol. 31, 520-530 (1995).
14. L. P. Whitney et al., Frontiers Microbiol. 2 (2011).
15. P. D. Chappell et al., ISME Journal 6, 1728-1739 (2012).
16. A. M. Hynes, Massachusetts Institute of Technology (MIT) (2009).
17. K. Suzuki et al., Biogeosci. 11, 2503-2517 (2014).
18. P. J. Keeling et al., Plos Biol. 12 (2014).
19. R. D. Groussman, M. S. Parker, E. V. Armbrust, PloS One 10 (2015).
20. J. Ashworth et al., Marine Genom. 26, 21-28 (2016).
21. J. Morrissey et al., Curr. Biol. 25, 364-371 (2015).



